Are you wondering where to find some very old proteins? If so, look no further than your nearest
fossilized ostrich eggshell in equatorial Africa, for it's there that protein preserved for 3.8 million years has been found. That’s over 5 times older than the 700,000 year old DNA found in the Arctic. This might seem strange given the higher, tropical temperatures in Africa, and the effect that time and temperature have on the degradation of biomolecules.
Addressing this conundrum, archaeologists (led by Prof Matthew Collins)
at the
University of York hypothesized that proteins could
survive intact if bound to a solid surface such as an eggshell. For example, within an ostrich's womb, '..several proteins are responsible for assembling the minerals that make up the ostrich eggshell. These proteins become trapped tightly within the mineral crystals themselves.'1 In the case of ostrich eggshells, this mineral is calcium carbonate - calcite - (CaCO3).
Researchers from universities across the world (e.g. Denmark, US, South Africa and the UK) used a combination of experimental and computational methods to determine how the eggshell protected biomolecules over millenia. Researchers from the University of Sheffield used computational classical molecular dynamics to probe the atomistic binding geometries between the calcite and protein (similar to this research.) They used the Collaborative Computational Project
5’s (CCP5) classical molecular dynamics program – DL_POLY – and discovered that the part of the protein that binds most strongly to the calcite surface of the eggshell, is most likely to remain preserved.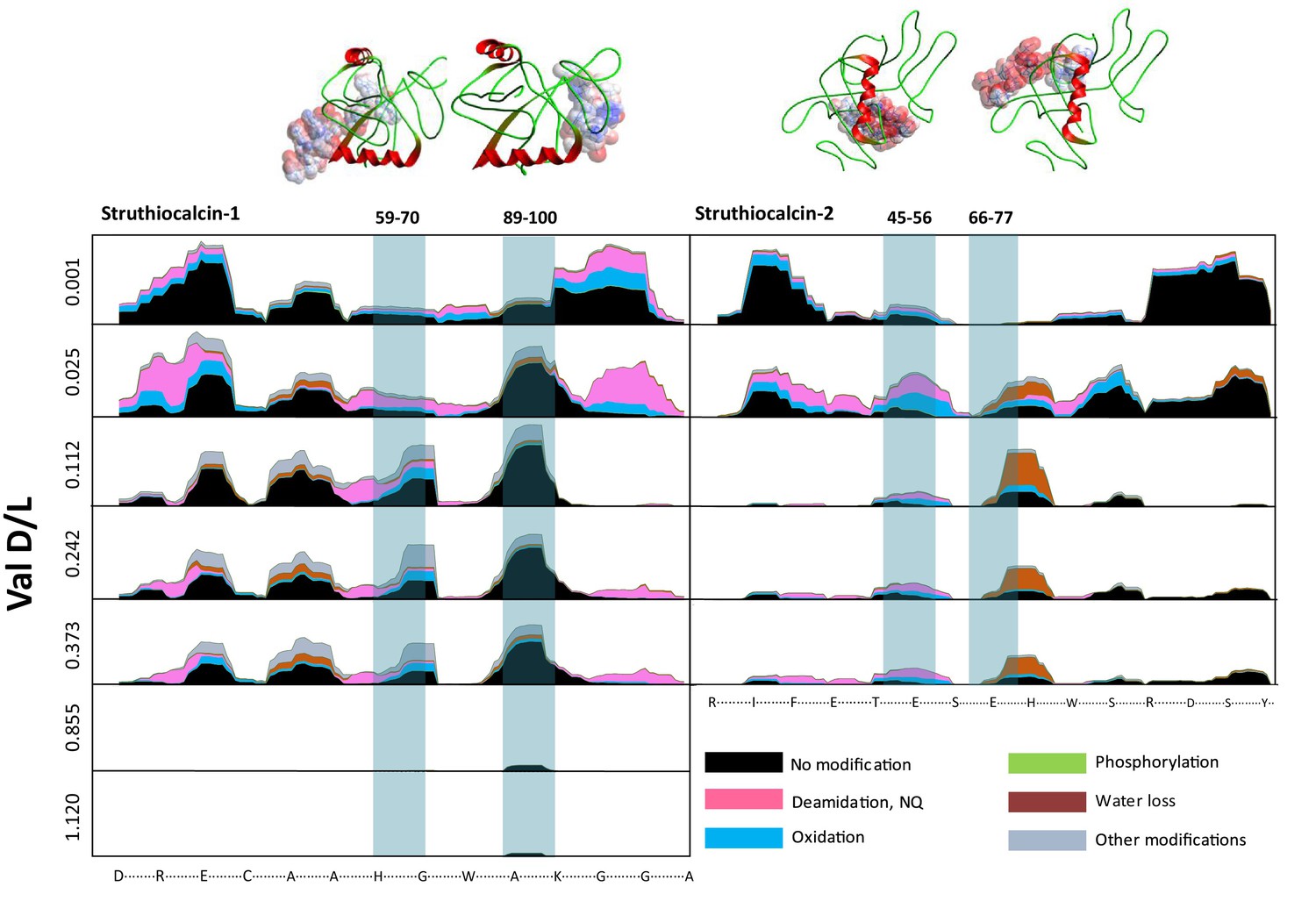

1 https://elifesciences.org/articles/17092#abstract Figure (from paper shown to the left) showing survival of protein constituents (i.e. peptides) with images from the DL_POLY simulations shown at the top
Although fragments of amino acids have
been found in much older fossils, whole protein sequences contain more valuable information, such as the function of the protein. This
can lead to robust evolutionary information
obtained from ‘biological barcodes’, an improvement on the previous need to
resort to inferring evolutionary relationships.
Atomistic modelling methods are powerful tools enabling us to explore the atomic-level interactions of a wide range of systems, and they work especially well in combination with experimental methods, as in the example given above. For further information about atomistic modelling carried out within the Chemistry Group at STFC, click here

Some of the material used in this case study was adapted from a BBC News & Environment article